Research
1. LightZymes: Artificial proteins that use light for catalysis
Within the ERC-starting grant project (starting 2018), we will create proteins that use light to catalyse regio- and stereoselective reactions. During evolution, new types of catalytic activities have been unlocked by utilizing additional cofactors. We will follow this thread of development and combine the strengths of bio- and photocatalysis: organo-photocatalysts will be placed as artificial cofactors into proteins to provide the protein with novel catalytic reactivities, and the proteins will be evolved to assist the catalytic reaction and to render it regio- and stereoselective.
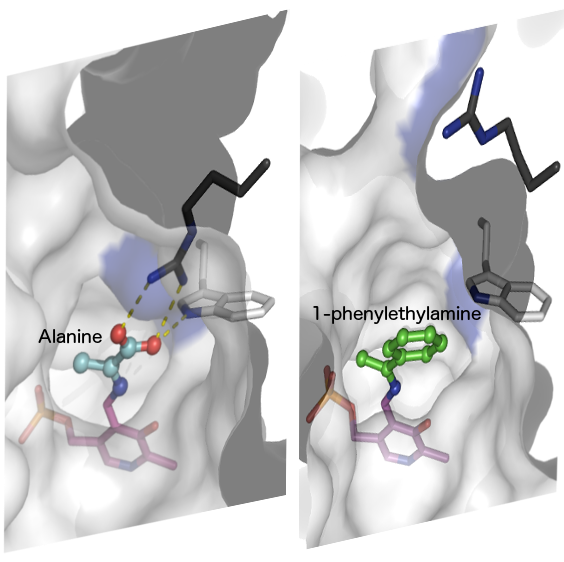
A) Structure – sequence – function relationships and enzyme discovery
Our aim is to relate general enzymatic properties like substrate specificity to patterns in protein sequence.
We demonstrated the power of this strategy for the in silico discovery of (R)-selective amine-transaminases for which no protein sequence was reported. By rational design we were able to identify a plausible ancestor and necessary key mutations, which we believed, should allow the conversion of (R)-amines. Instead of constructing the predicted mutants in the lab, 20 proteins could be identified in protein databases that already carry these mutations. Indeed, 15 enzymes converted (R)-amines significantly (Nat Chem Biol, 2010) and five enzymes turned out to be highly useful for the asymmetric synthesis of a panel of different (R)-amines (Adv Synth Catal 2011).
We also identified the catalytic function of six proteins with solved crystal structures (deposited in the protein data bank, but biochemically uncharacterized) and their biocatalytic potential as transaminases and imine reductases (ChemCatChem 2013, Appl Microb Biotechnol 2016, JMCB 2014).
From the transaminase structures, we elucidated a molecular model how dual substrate recognition works (click here for an animation)
By a comprehensive bioinformatic analysis, we revealed important active site residues that determine substrate and reaction specificity of 28 different enzymes belonging to PLP fold class I (Biotech Adv 2015).
B) Protein engineering and application of discovered enzymes
Our current focus is the biocatalytic synthesis of functionalized and beta chiral amines (builiding blocks/pharmaceuticals) using transaminases (Adv Synth Catal 2015) and imine reductases. Especially imine reductases are attractive, as they allow the asymmetric one-step synthesis of secondary or tertiary amines (ChemBioChem 2017) from ketones. We synthesized both enantiomers of rasagiline using stereo complementary imine reductases (Green Chem 2017). To explore and tailor the substrate specificity, we employ methods of protein engineering, e.g. to relax the cofactor preference of an imine reductases so that NADH can be used in biocatalysis instead of NADPH (J Biotechnol 2016).

