Arbeitskreis "Synthetische & Strukturelle Biochemie"
Lehrstuhlinhaber
Prof. Dr. rer. nat. Michael Lammers
Institut für Biochemie
Arbeitskreis Sythetische und strukturelle Biochemie
Felix-Hausdorff-Str. 4
17487 Greifswald
Telefon +49 3834 420-4356
michael.lammers@uni-greifswald.de
Kontakt
Kathrin Krassow
Sekretariat
Institut für Biochemie
Arbeitskreis Molekulare Strukturbiologie
Felix-Hausdorff-Str. 4
17489 Greifswald
Telefon +49 3834 420-4391
Telefax +49 3834 420-4373
kathrin.krassowuni-greifswaldde
Lehrstuhlinhaber
Prof. Dr. rer. nat. Michael Lammers
Sekretariat
Kathrin Krassow
Institut für Biochemie
Synthetische & Strukturelle Biochemie
Felix-Hausdorff-Str. 4
17487 Greifswald
Telefon +49 3834 420-4391
kathrin.krassow@uni-greifswald.de
Informationen für Studierende finden Sie unter Lehre zu Ihrem jeweiligen Studiengang.
HEC25 Heart of European Biocrystallography Meeting
The HEC25 was taking place from 21 - 23 Sept 2023 in Salem (MV), directly located at the Kummerower See with fantastic talks and lively discussions around applications of crystallography in the fields of biochemistry, medicine, microbiology, etc.
A huge 'Thank You' goes out to all attendees for making this years HEC such a memorable experience! :)
Forschungsschwerpunkte
Lysine acetylation was discovered in 1960s by Vincent Allfrey and colleagues to occur on histones to regulate RNA synthesis. Nearly three decades later the yeast protein Sir2 was shown to affect longevity in budding yeast. This was a remarkable finding as it shows that a single protein is enough to regulate lifespan of a whole organism. Later it was found that Sir2 has an NAD+-dependent deacetylase activity. Sirtuins have been shown later to be involved in lifespan regulation also in flies, worms and mice. Furthermore, they affect healthy aging and do play protective roles in the development of severe diseases such as neurodegenerative diseases as well as cancer. Therefore, the lysine acetylation status in the proteome is very likely contributing to disease development.
The identification of diverse charged and uncharged lysine acylations puts another level of complexity on this post-translational modification. Thousands of lysine acetylation sites, and of other acylations occuring at lysine side chains, have been found in the proteome of diverse organisms by quantitative mass-spectrometry. One of the major challenges in the acylation research field is to distinguish between biologically relevant and physiologically unimportant sites. Notably, less than 1% of these lysine acylation events were functionally characterised so far.
We use a combined synthetic biological, biophysical and cell biological approach to structurally and functionally investigate how protein function is regulated by site-specific lysine acetylation, and by other acylations, and how a dysfunction in this post-translational machinery contributes to disease development. Furthermore, we examine how lysine acylation is regulated by sirtuins and lysine-acetyltransferases and how these enzymes target their substrates to unravel molecular determinants of substrate specificity. These studies will give valuable information on the regulation of lysine acylation in vivo and furthermore will support the development of isoform specific activators or inhibitors for KDAC/sirtuin/KAT function for therapeutic applications.
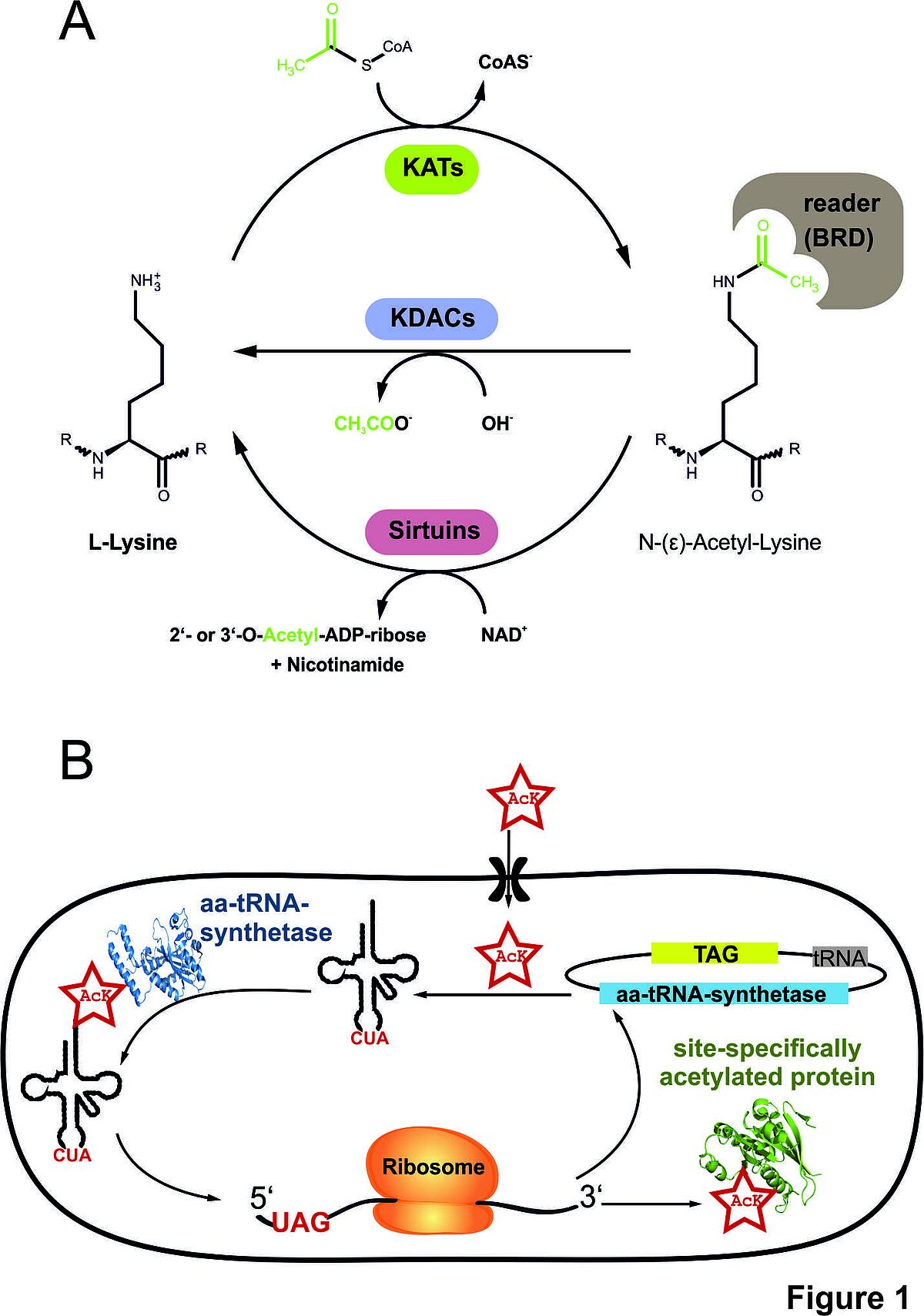
Figure 1: (A) Lysine-acetylation/acylation is a dynamically regulated post-translational modification catalysed by writers, lysine-acetyltransferases (KATs), and erasers, lysine-deacetylases (KDACs or sirtuins). Acetylated lysine side chains can be targeted by readers, bromodomain (BRD) containing proteins (B) and The genetic-code expansion concept (GCEC). We use a synthetically evolved acetyl-lysyl-tRNA-synthetase/tRNACUA pair derived from PylRS/pylT from Methanosarcina barkeri to site-specifically incorporate N-(ε)-acetyl-L-lysine into proteins. In the future, we want to expand our studies also to other lysine acylations.
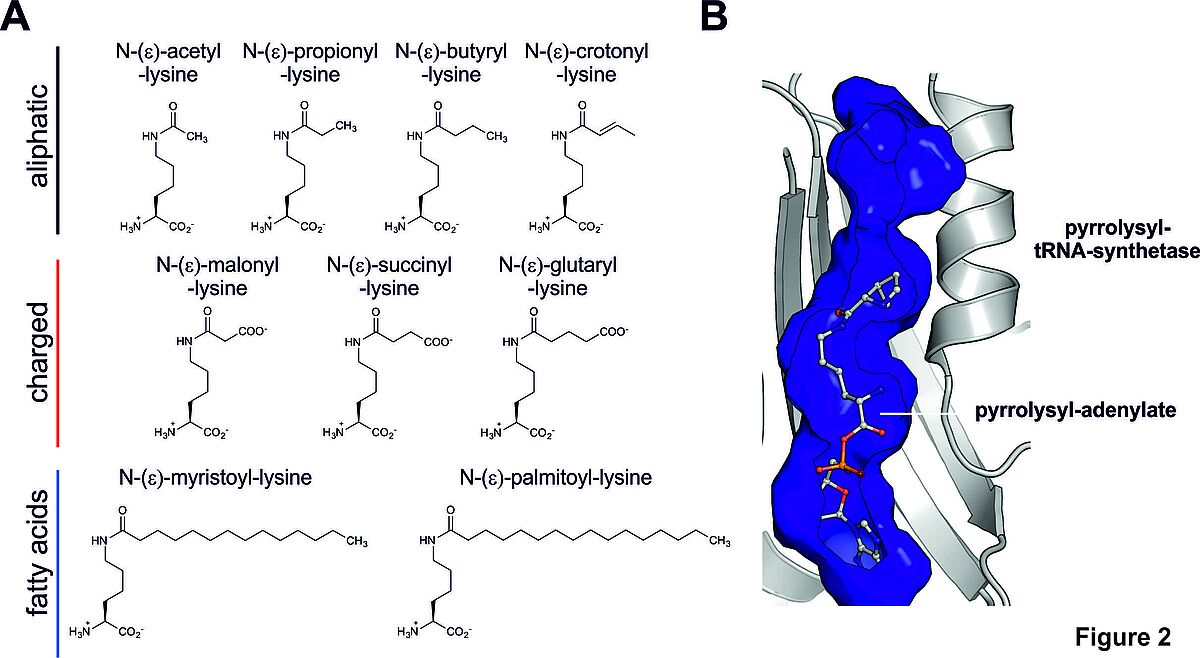
Figure 2: Lysine side chains can be targeted by a variety of different lysine-acylations, including aliphatic, charged and uncharged acylations. For many acylations a rigorous functional characterisation has not been performed so far. (B) We use a PylRS, the pyrrolysyl-tRNA-synthetase from Methanosarcina barkeri, as a synthetic-biological tool to incorporate lysine-acylations site-specifically into proteins.
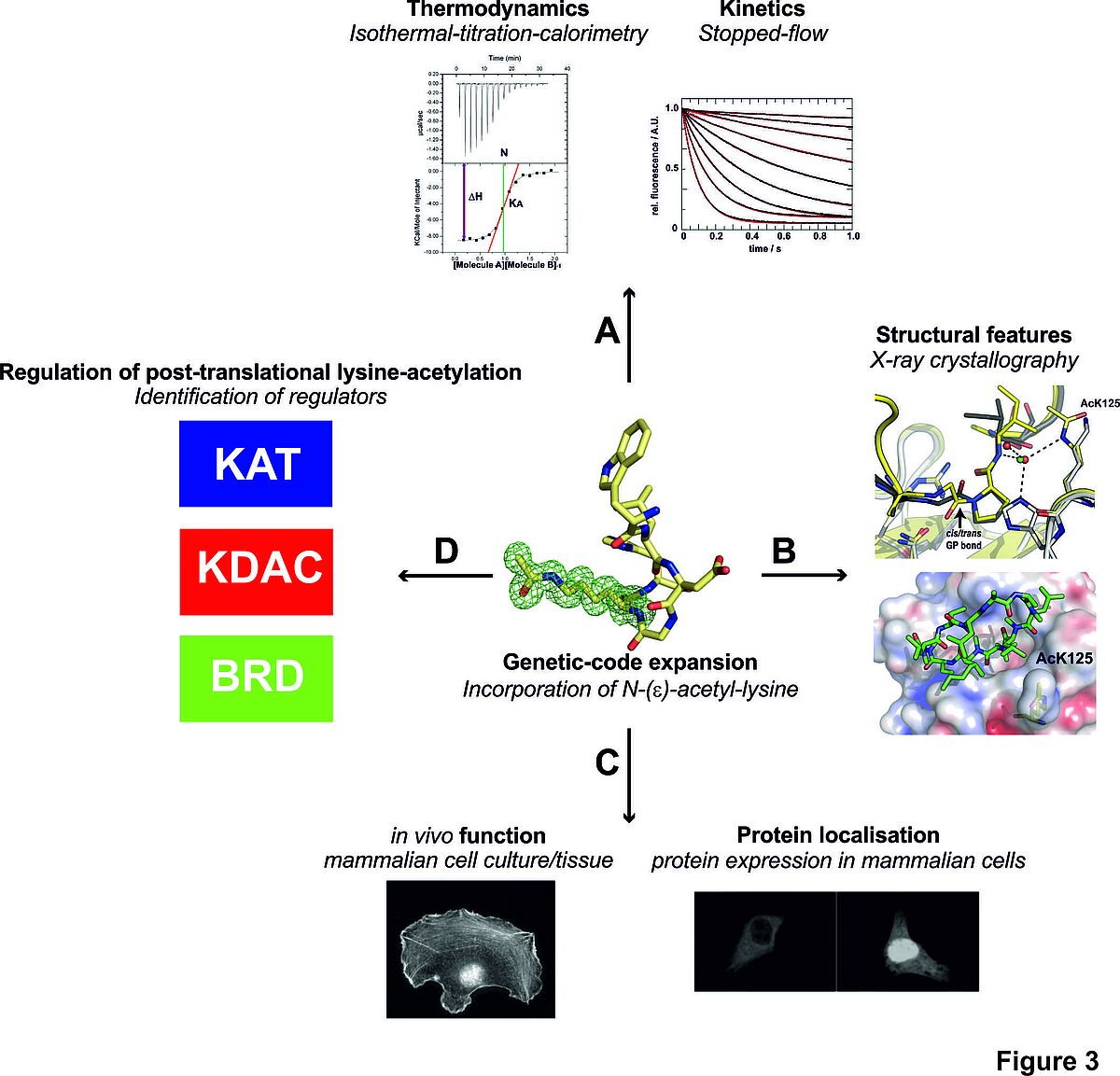
Figure 3: Structural and functional studies of the effect of lysine-acetylation on protein function. (A) We study the influence of lysine-acylation on the thermodynamics and kinetics of protein-protein interactions and on the protein-structure (B). (C) To disctiminate between biologically important and unimportant sites, we study the impact of lysine-acetylation in mammalian cells/tissues (D) and examine how it is regulated by KDACs, KATs and how it is targeted by BRD-containing proteins.